

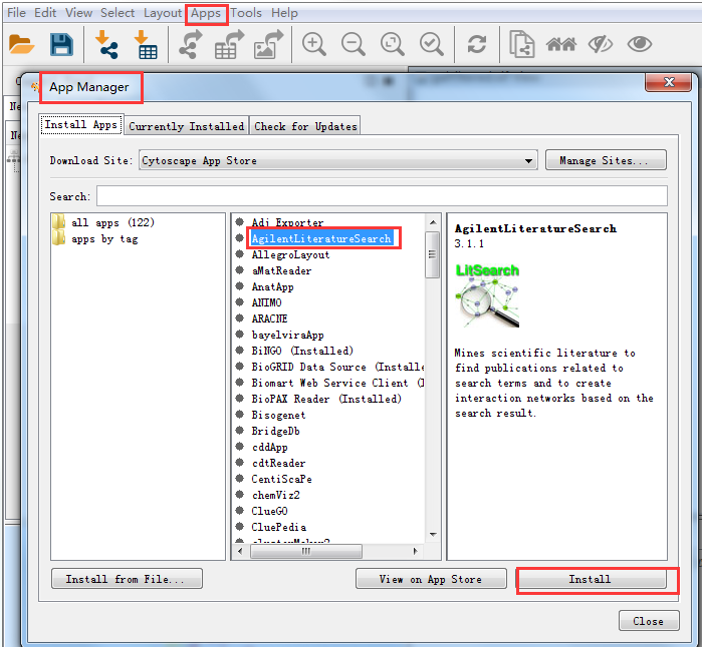
UCSF clusterMaker is a Cytoscape plugin that unifiesĭifferent clustering techniques and displays into a single In this screenshot, theĮxpression data in the sampleData file galFiltered.cys has been clustered using the hierarchical methodĪnd displayed as a heatmap with associated dendrogram. it might join two nodes that were NOT meant to be joined).Figure 1.

Though, I'd love to see arguments in favour of pydot.Įdit1: if you want to plot by attributed and not by label, see this answer: NetworkX node attribute drawing note that relabeling the way I suggested does not always have the intended semantics (e.g. Though I want to leave this link about pydot visualization since it seems very useful in general: Display graph without saving using pydot and probably provides the pydot answer for others if they need it. graph ( (, ), num_nodes = 6 ) print ( f' \n'
CYTOSCAPE TUTORIAL CODE
Also, networkx is nice because I can transform dgl graphs to networkx too (and the relabeling was simple).Ĭonsidering those reasons let me give you the example code I wrote that does what you want using pygraphviz (but you could do it with pydot if you figured out how, transforming to pydot obj is trivial using networkx see my previous link):
It's hard to make these decisions but I can't stay on this forever and this seems mindful enough.
CYTOSCAPE TUTORIAL HOW TO
Pygravix also has docs which although small make me happy (though not as good as graphviz but idk how to make graphviz graphs from networkx). In addition, I don't know how to convert a networkx directly to a graphviz obj (since graphviz has the best docs and highest user base ~19k so I did prefer that), so I will go with pygraphviz for those reasons. Īlso it seems that pygraphviz has more granular control than regular graphviz ref: Graphviz vs PyGraphViz. Idk if it's that it doesn't look nice on my browser or that it makes me feel that project is not being taken seriously by it's developers idk something just doesn't feel right even if it has a higher set of users pydot 15k vs pygraphviz 4k) reference. In addition, pydot as of now does not really have docs (which personally bothers me. Much of the NetworkX tutorial at is applicable to PyGraphviz. The API is very similar to that of NetworkX. It seems from the docs in pygraphviz that it has a similar api so it might be the easiest to use if you already want like to use networkx ( ): My suspicion is that pydot and pygraphviz are the best for some reason since networkx only supports those two.
CYTOSCAPE TUTORIAL SOFTWARE
Thus my suggestion is to transform the graph to some format that has dedicated software for graph visualization and then draw (e.g. For example, Cytoscape can read the GraphML format, and so, networkx.write_graphml(G, path) might be an appropriate choice. To use these and other such tools, you should export your NetworkX graph into a format that can be read by those tools. Notable examples of dedicated and fully-featured graph visualization tools are Cytoscape, Gephi, Graphviz and, for LaTeX typesetting, PGF/TikZ. Proper graph visualization is hard, and we highly recommend that people visualize their graphs with tools dedicated to that task. In the future, graph visualization functionality may be removed from NetworkX or only available as an add-on package. NetworkX provides basic functionality for visualizing graphs, but its main goal is to enable graph analysis rather than perform graph visualization. They explicitly warn you that graph visualization is hard and networkx is mainly meant for graph analysis (from ): I feel a better answer is not to use networkx to draw.


 0 kommentar(er)
0 kommentar(er)
